Identification of novel targets for molecular imaging
Fluorescence molecular imaging using a targeted tumor-specific marker that is selected based on its expression in tumors can be used to identify malignancy or visualize treatment response. These biomarkers are preferably selectively overexpressed in dysplastic or malignant cells. Naturally, these targets have to be identified first. Within the OMIG we identify novel targets for a wide variety of clinical end-goals.
Functional Genomic mRNA profiling
A study performed within our group by Hartmans et al.,1 identified novel imaging targets for the detection of early colorectal lesions with biostatistics using functional genomic mRNA (FGmRNA) profiling for the first time. Early detection and radical removal of lesions improves the life expectancy of the patients but is challenging due to non-specific tissue morphology. In standard white-light endoscopy, for example, there is a detection miss rate of up to 25%.2 In particular flat and small lesions are difficult to detect. Enhancing the regular endoscopic techniques with molecular imaging using a ‘red flag’ strategy to localize the lesions can improve the optical discrimination of these (dysplastic) lesions.
To identify relevant target proteins, DNA changes that occur in adenomas, and influence the protein expression levels of cells, can be addressed. To enable identification of target proteins that distinguish (dysplastic) lesions, FGmRNA profiling was introduced. Using this technique gene expression data for major non-genetic factors is corrected, which enhances the view on the downstream effect of genomic alterations occurring on the downstream effect resulting in an altered gene expression level.3 This technique is based on principal component analysis performed on expression profiles of publicly available samples.
Eventually, based on twenty-six normal mucosa and forty-seven adenoma samples, 4524 genes were found to show overexpression in adenoma samples. These genes were ranked to their level of significance and their appearance on the cell, as can be seen in the Figure below.
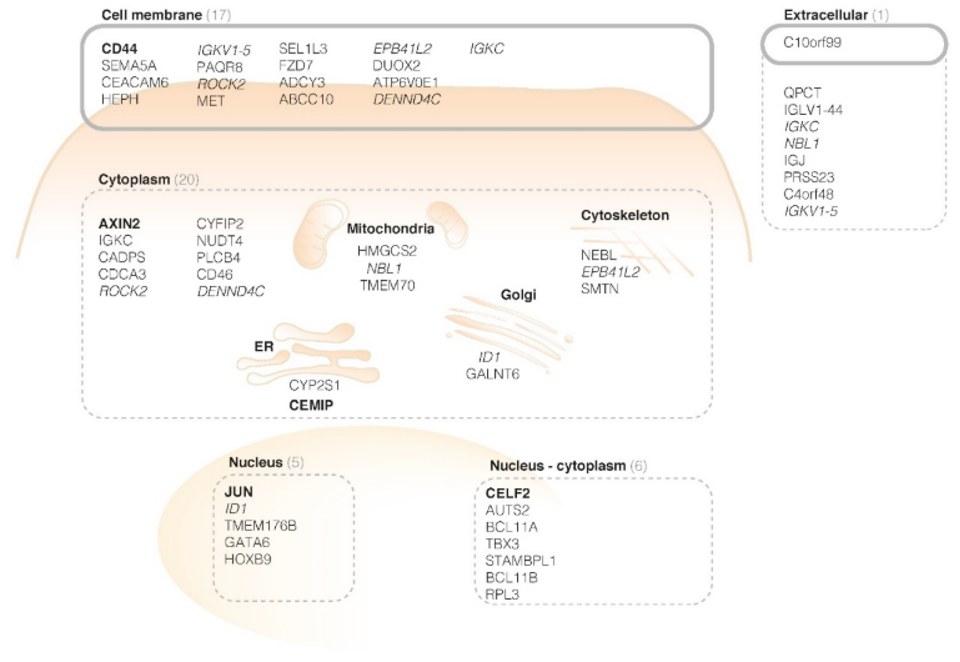
CD44v6 was found to be a promising target for the detection of early colorectal lesions. To confirm the hypothesis, a next step was immunohistochemistry. The presence of CD44v6 in colorectal adenomas was seen in human tissue. Secondly, the membranous overexpression of CD44v6 was shown in adenomatous mice crypts with fluorescence imaging experiments.
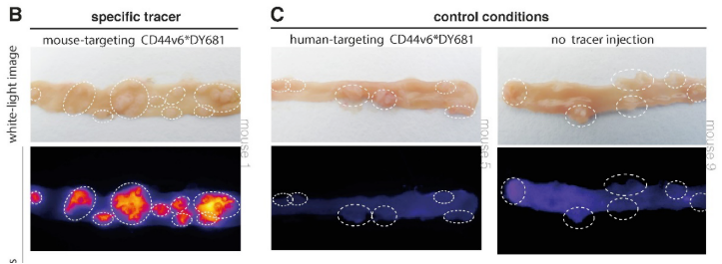
Based on this structured research using bioinformatics and a functional approach, CD44v6 is identified as a promising target for ‘red flag’ adenoma detection and suitable for clinical translation.
Independent Component Analysis
Hartmans et al. base their hypothesis on FGmRNA but hypotheses can also be based on other aspects. For example, literature or independent component analysis (ICA). With ICA, expression patterns of tumor biopsies can be dissected into statistically independent transcriptional components (TC) of single biological processes. This can be focused on, for example, disease free survival, or, to identify markers for molecular imaging, certain metastases patterns. In each TC the most important genes and therewith expression profiles can be identified. The ICA approach provides us hypotheses, just like FGmRNA, which will always structurally need to be confirmed using IHC, ex vivo fluorescence imaging and eventually in an in vivo clinical trial.
Bibliography
-
Hartmans E, Orian-Rousseau V, Matzke-Ogi A, et al. Functional genomic mRNA profiling of colorectal adenomas: Identification and in vivo validation of CD44 and splice variant CD44v6 as molecular imaging targets. Theranostics. 2017;7(2):482-492. doi:10.7150/thno.16816
-
van Rijn JC, Reitsma JB, Stoker J, Bossuyt P, van Deventer SJ, Dekker E. Polyp Miss Rate Determined by Tandem Colonoscopy: A Systemat… : Official journal of the American College of Gastroenterology | ACG. Am J Gastroenterol. 2006;101(2):343-350. Accessed May 18, 2022. https://journals.lww.com/ajg/Abstract/2006/02000/Polyp_Miss_Rate_Determined_by_Tandem_Colonoscopy_.25.aspx
-
N Fehrmann RS, Karjalainen JM, Krajewska M, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet Vol. 2015;47(2). doi:10.1038/ng.3173
